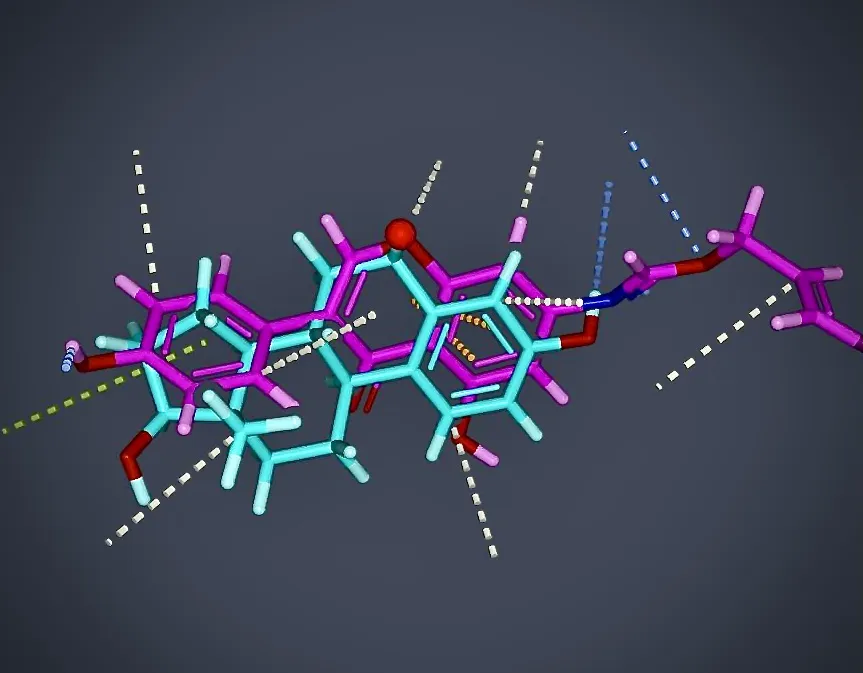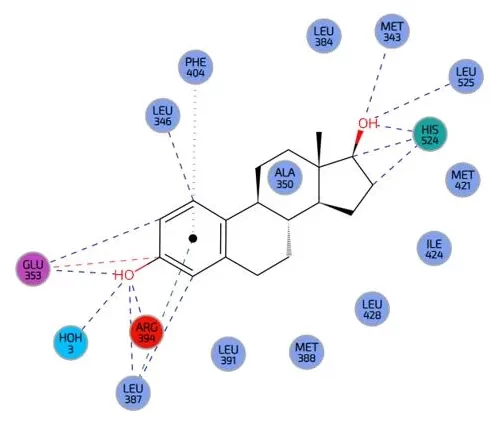
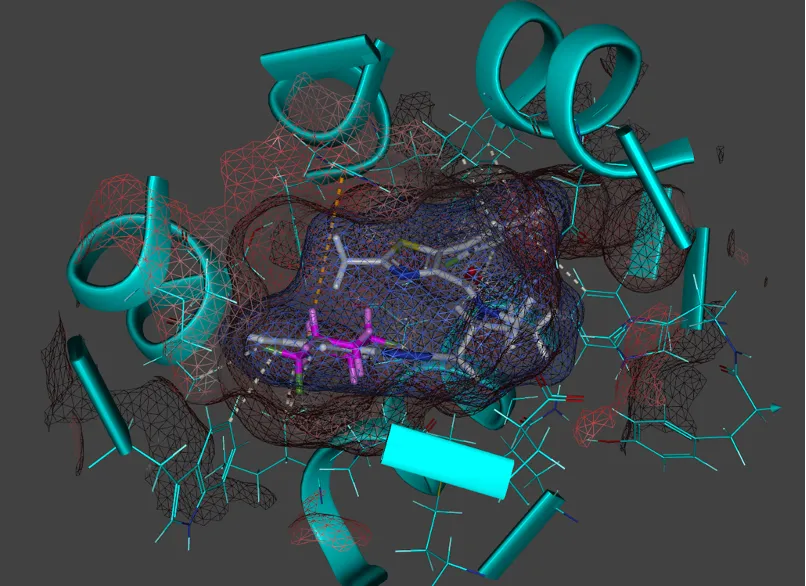
About molecular docking
Molecular docking is a computational technique (in silico) used to predict how small molecules (ligands) interact with a target protein, aiding in food product development, assessment of food safety, drug discovery, designing host-guest chemistries and predicting complexation, for example, of metal ions. By simulating these interactions in silico, researchers can identify potential food ingredient compound or drug candidates, and understand their binding affinities in the early stages of a project, which is crucial in the food product regulatory approval or drug discovery process.
Our team utilises state-of-the-art software and algorithms to ensure accurate and reliable results. Emerging technologies, such as AI agents and tools, as well as virtual reality (VR) headsets, allow researchers to visualise and manipulate protein-ligand complexes in 3D with a unique perspective, providing unique insights into binding interactions, structural flexibility, and food compound or drug optimisation, enhancing rational food product and drug design strategies.
ISMD Molecular Docking Services
Our molecular docking services are designed to assist researchers, food, herbal and pharmaceutical companies in:
Protein-Ligand Docking: Predicting interactions between small molecules and proteins.
Understanding Molecular Interactions: Gaining insights into the binding mechanisms between ligands and their targets., for instance, for safety assessments.
Homology-docking studies: Enabling insights and predicting protein structures when experimental data is unavailable.
Virtual complexation docking experiments: Analysing in silico protein-ligand interactions to assess food and herbal compound safety or optimise drug design.
Contact us